Spatial Transcriptome: A Novel Technology is Now Available at NGS Core
Dear Professors,
We recently adopted a novel and very cool technology – spatial transcriptome. It can be applied to any tissue cryosections for finding regions/cells/cell layers with particular gene expression and the corresponding marker gene for studies such as cardiac defects, neuronal development, cancer progression, and evo devo, etc.
Please follow the link below for more details.
https://10xgenomics.com/products/spatial-gene-expression/
Please kindly contact us if you are interested, and welcome to help spread the news!
Sincerely,
NGS High Throughput Genomics Core
Biodiversity Research Center, Academia Sinica
Launching of the 3rd-Gen sequencing service of PacBio Sequel system
Dear Professors,
We are happy to announce the launching of the 3rd-Gen sequencing service of PacBio Sequel system.
Built on Single Molecule, Real-Time (SMRT) technology, the scalable, high-throughput Sequel System delivers long-read sequencing, uniform coverage, and high consensus accuracy, and provides greater data output than the RS II system for its much greater capacity of the sequencing cell.
Please kindly contact us if you are interested, and welcome to help spread the news!
Best regards,
NGS High Throughput Genomics Core
Biodiversity Research Center, Academia Sinica
PacBio – Sequel
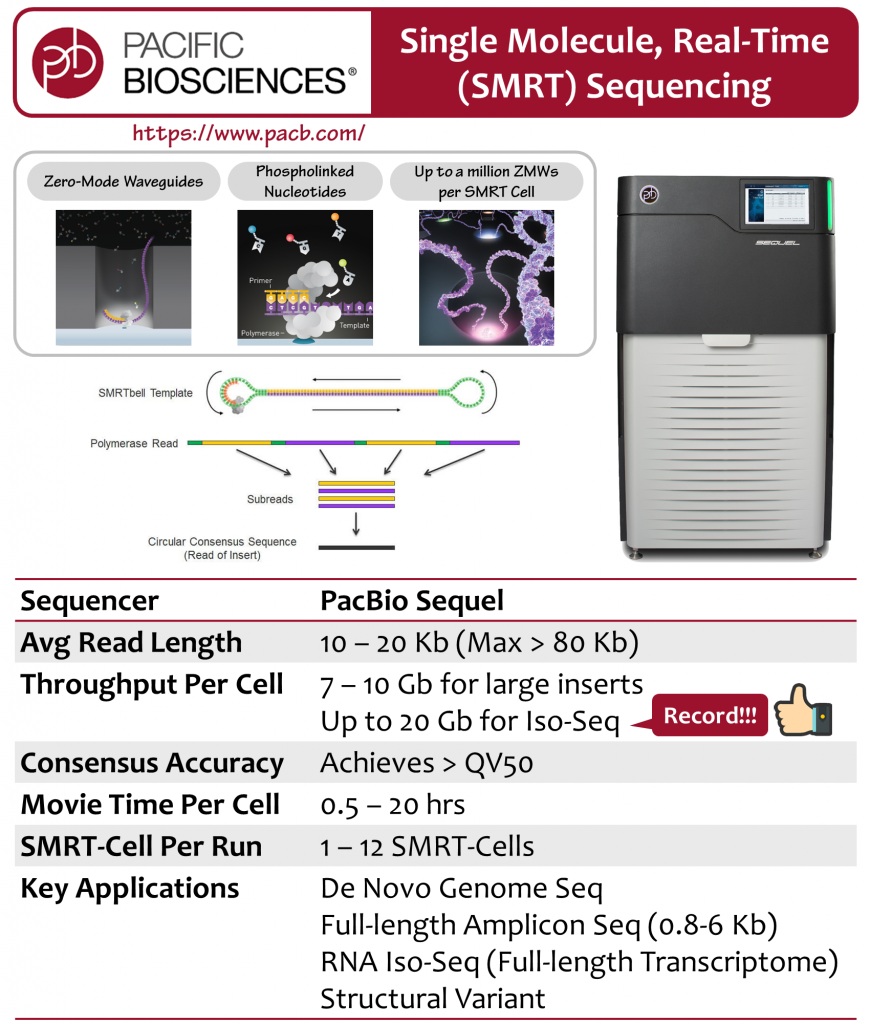
With SMRT (Single Molecule Real-Time) technology, PacBio Sequel enables the real-time detection of DNA synthesis, which is done by incorporation of phospholinked nucleotides, and allows DNA polymerase to complete natural process of elongation. The events of DNA synthesis occurs in the bottom of ZMWs (zero-mode waveguides), allowing the illumination to be identified from background noises such as unincorporated free nucleotides.
PacBio Sequel system provides long-read sequencing, which facilitates assemblies of difficult genomic regions. It also offers benefits such as uniform coverage and high consensus accuracy, and becomes a powerful technology for high quality de novo genome study, full-length transcriptome investigation, haplotype phasing, and long length amplicon research.