Please support us by sharing your publications. Thank you!
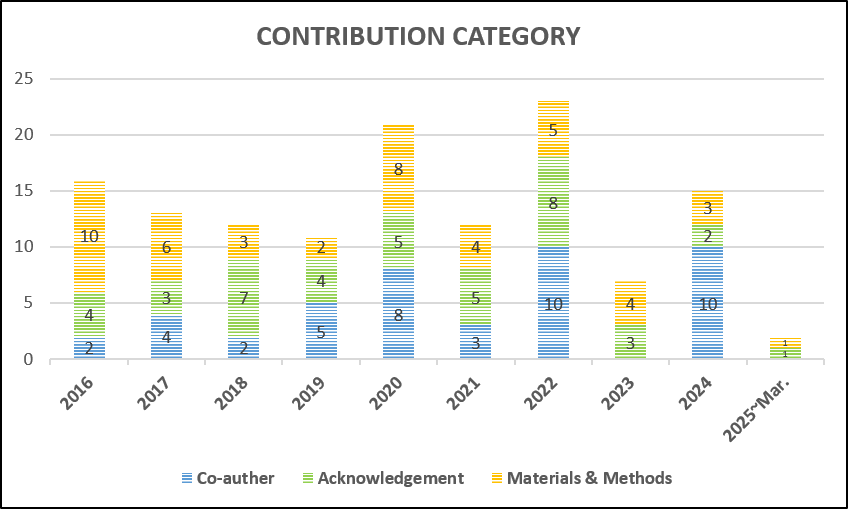
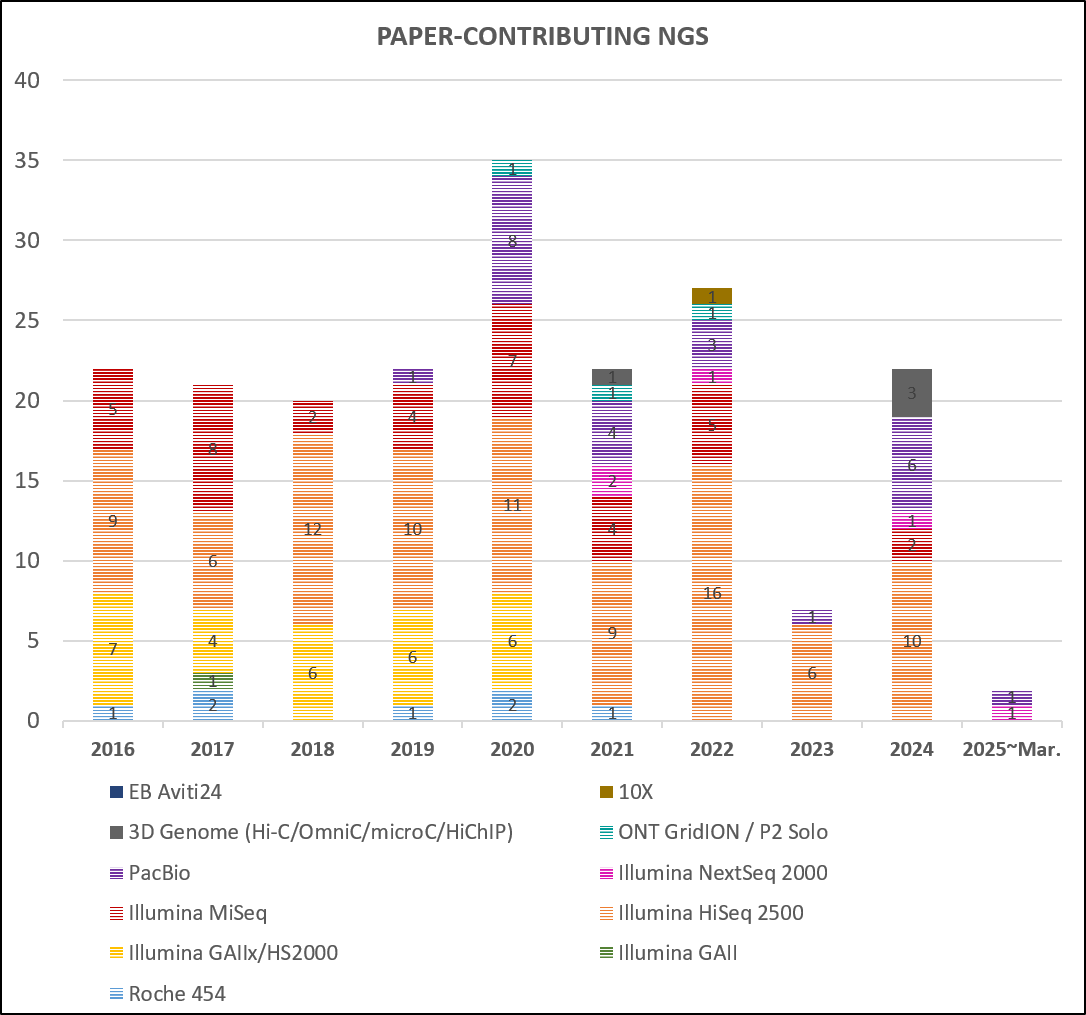
If you use data from our sequencing services in your publication, we’d really appreciate it if you could mention the NGS core facility in the acknowledgements.
2025
Wang, P.H., Wu, T.Y., Chen, Y.L., Gicana, R.G., Lee, T.H., Chen, M.J., Hsiao, T.H., Lu, M.J., Lai, Y.L., Wang, T.Y., et al. (2025).
Bacterial estrogenesis without oxygen: Wood-Ljungdahl pathway likely contributed to the emergence of estrogens in the biosphere.
Proc Natl Acad Sci U S A 122, e2422930122, 10.1073/pnas.2422930122.
https://www.ncbi.nlm.nih.gov/pubmed/40053361
Parada, C.M., Yan, C.S., Hung, C.Y., Tu, I.P., Hsu, C.P., and Shih, Y.L. (2025).
Growth-dependent concentration gradient of the oscillating Min system in Escherichia coli.
J Cell Biol 224, 10.1083/jcb.202406107.
https://www.ncbi.nlm.nih.gov/pubmed/39621132.
Bacterial estrogenesis without oxygen: Wood-Ljungdahl pathway likely contributed to the emergence of estrogens in the biosphere.
Proc Natl Acad Sci U S A 122, e2422930122, 10.1073/pnas.2422930122.
https://www.ncbi.nlm.nih.gov/pubmed/40053361
Growth-dependent concentration gradient of the oscillating Min system in Escherichia coli.
J Cell Biol 224, 10.1083/jcb.202406107.
https://www.ncbi.nlm.nih.gov/pubmed/39621132.
2024
| Wang, P.-H., Wu, T.-Y., Chen, Y.-L., Gicana, R.G., Lee, T.-H., Chen, M.-J., Hsiao, T.-H., Jade Lu, M.-Y., Lai, Y.-L., Wang, T.-Y., et al. (2024). Aromatase-independent estrogenesis: Wood-Ljungdahl pathway likely contributed to the emergence of estrogens in the biosphere. bioRxiv, 2024.2010.2024.619986, 10.1101/2024.10.24.619986. https://www.biorxiv.org/content/biorxiv/early/2024/10/24/2024.10.24.619986 |
| Le, M.H., Morgan, B., Lu, M.Y., Moctezuma, V., Burgos, O., and Huang, J.P. (2024). The genomes of Hercules beetles reveal putative adaptive loci and distinct demographic histories in pristine North American forests. Mol Ecol Resour 24, e13908, DOI: 10.1111/1755-0998.13908. https://www.ncbi.nlm.nih.gov/pubmed/38063363. |
| Mattos, F.M.G., Dreyer, N., Fong, C.L., Wen, Y.V., Jain, D., De Vivo, M., Huang, Y.S., Mwihaki, J.K., Wang, T.Y., Ho, M.J., et al. (2024). Potential PCR amplification bias in identifying complex ecological patterns: Higher species compositional homogeneity revealed in smaller-size coral reef zooplankton by metatranscriptomics. Mol Ecol Resour 24, e13911, DOI: 10.1111/1755-0998.13911. https://www.ncbi.nlm.nih.gov/pubmed/38063371. |
| Kao, H.J., Lin, T.Y., Hsieh, F.J., Chien, J.Y., Yeh, E.C., Lin, W.J., Chen, Y.H., Ding, K.H., Yang, Y., Chi, S.C., et al. (2024). Highly efficient capture approach for the identification of diverse inherited retinal disorders. NPJ Genom Med 9, 4, DOI: 10.1038/s41525-023-00388-3. https://www.ncbi.nlm.nih.gov/pubmed/38195571. |
| Kao, H.J., Lin, T.Y., Hsieh, F.J., Chien, J.Y., Yeh, E.C., Lin, W.J., Chen, Y.H., Ding, K.H., Yang, Y., Chi, S.C., et al. (2024). Highly efficient capture approach for the identification of diverse inherited retinal disorders. NPJ Genom Med 9, 4, DOI: 10.1038/s41525-023-00388-3. https://www.ncbi.nlm.nih.gov/pubmed/38195571. |
| Hung, Y.L., Hong, S.F., Wei, W.L., Cheng, S., Yu, J.Z., Tjita, V., Yong, Q.Y., Nishihama, R., Kohchi, T., Bowman, J., et al. (2024). Dual regulation of cytochrome P450 gene expression by two distinct small RNAs, a novel tasiRNA and miRNA, in Marchantia polymorpha. Plant Cell Physiol, DOI: 10.1093/pcp/pcae029. https://www.ncbi.nlm.nih.gov/pubmed/38545690. |
| Lin, M.D., Chuang, C.H., Kao, C.H., Chen, S.H., Wang, S.C., Hsieh, P.H., Chen, G.Y., Mao, C.C., Li, J.Y., Jade Lu, M.Y., and Lin, C.Y. (2024). Decoding the genome of bloodsucking midge Forcipomyia taiwana (Diptera: Ceratopogonidae): Insights into odorant receptor expansion. Insect Biochem Mol Biol 168, 104115, DOI: 10.1016/j.ibmb.2024.104115. https://www.ncbi.nlm.nih.gov/pubmed/38570118. |
| Chan, Y.F., Lu, C.W., Kuo, H.C., and Hung, C.M. (2024). A chromosome-level genome assembly of the Asian house martin implies potential genes associated with the feathered-foot trait. G3 (Bethesda) 14, DOI: 10.1093/g3journal/jkae077. https://www.ncbi.nlm.nih.gov/pubmed/38607414. |
| Chang, Y.H., Hsu, M.F., Chen, W.N., Wu, M.H., Kong, W.L., Lu, M.J., Huang, C.H., Chang, F.J., Chang, L.Y., Tsai, H.Y., et al. (2024). Functional and structural investigation of a broadly neutralizing SARS-CoV-2 antibody. JCI Insight 9, DOI: 10.1172/jci.insight.179726. https://www.ncbi.nlm.nih.gov/pubmed/38775156. |
| Hong, S.F., Wei, W.L., Pan, Z.J., Yu, J.Z., Cheng, S., Hung, Y.L., Tjita, V., Wang, H.C., Komatsu, A., Nishihama, R., et al. (2024). Molecular Insights into MpAGO1 and Its Regulatory miRNA, miR11707, in the High-Temperature Acclimation of Marchantia polymorpha. Plant Cell Physiol 65, 1414-1433, 10.1093/pcp/pcae080. https://www.ncbi.nlm.nih.gov/pubmed/38988198. |
| Reynaud, M., Vianello, S., Lee, S.-H., Salis, P., Wu, K., Frederich, B., Lecchini, D., Besseau, L., Roux, N., and Laudet, V. (2024). The multi-level effect of chlorpyrifos during clownfish metamorphosis. bioRxiv, 2024.2008.2002.606305, 10.1101/2024.08.02.606305. http://biorxiv.org/content/early/2024/09/27/2024.08.02.606305.abstract. |
| Chou, C., Lin, C.Y., Lin, C.Y., Wang, A., Fan, T.P., Wang, K.T., Yu, J.K., and Su, Y.H. (2024). Tracing the evolutionary origin of chordate somites in the hemichordate Ptychodera flava. Integr Comp Biol, 10.1093/icb/icae020. https://www.ncbi.nlm.nih.gov/pubmed/38637301. |
| Lin, C.Y., Marletaz, F., Perez-Posada, A., Martinez-Garcia, P.M., Schloissnig, S., Peluso, P., Conception, G.T., Bump, P., Chen, Y.C., Chou, C., et al. (2024). Chromosome-level genome assemblies of 2 hemichordates provide new insights into deuterostome origin and chromosome evolution. Plos Biol 22, e3002661, 10.1371/journal.pbio.3002661. https://www.ncbi.nlm.nih.gov/pubmed/38829909. |
| Su, P.A., Chang, C.H., Yen, S.B., Wu, H.Y., Tung, W.J., Hu, Y.P., Chen, Y.I., Lin, M.H., Shih, C., Chen, P.J., and Tsai, K. (2024). Epitranscriptomic cytidine methylation of the hepatitis B viral RNA is essential for viral reverse transcription and particle production. Proc Natl Acad Sci U S A 121, e2400378121, 10.1073/pnas.2400378121. https://www.ncbi.nlm.nih.gov/pubmed/38830096. |
| Lin, M.D., Chuang, C.H., Kao, C.H., Chen, S.H., Wang, S.C., Hsieh, P.H., Chen, G.Y., Mao, C.C., Li, J.Y., Jade Lu, M.Y., and Lin, C.Y. (2024). Decoding the genome of bloodsucking midge Forcipomyia taiwana (Diptera: Ceratopogonidae): Insights into odorant receptor expansion. Insect Biochem Mol Biol 168, 104115, 10.1016/j.ibmb.2024.104115. https://www.ncbi.nlm.nih.gov/pubmed/38570118. |
2023
| Lu, C.W., Huang, S.T., Cheng, S.J., Lin, C.T., Hsu, Y.C., Yao, C.T., Dong, F., Hung, C.M., and Kuo, H.C. (2023). Genomic architecture underlying morphological and physiological adaptation to high elevation in a songbird. Mol Ecol 32, 2234-2251, DOI: 10.1111/mec.16875. https://www.ncbi.nlm.nih.gov/pubmed/36748940. |
| De Vivo, M., Chen, W.Y., and Huang, J.P. (2023). Testing the efficacy of different molecular tools for parasite conservation genetics: a case study using horsehair worms (Phylum: Nematomorpha). Parasitology 150, 842-851, DOI: 10.1017/S0031182023000641. https://www.ncbi.nlm.nih.gov/pubmed/37415562. |
| Negi, A., Liao, B.Y., and Yeh, S.D. (2023). Long-read-based Genome Assembly of Drosophila gunungcola Reveals Fewer Chemosensory Genes in Flower-breeding Species. Genome Biol Evol 15, DOI: 10.1093/gbe/evad048. https://www.ncbi.nlm.nih.gov/pubmed/36930539. |
| Wei, K.H., Lin, I.T., Chowdhury, K., Lim, K.L., Liu, K.T., Ko, T.M., Chang, Y.M., Yang, K.C., and Lai, S.B. (2023). Comparative single-cell profiling reveals distinct cardiac resident macrophages essential for zebrafish heart regeneration. Elife 12, DOI: 10.7554/eLife.84679. https://www.ncbi.nlm.nih.gov/pubmed/37498060. |
| Kuo, H.C., Yao, C.T., Liao, B.Y., Weng, M.P., Dong, F., Hsu, Y.C., and Hung, C.M. (2023). Weak gene-gene interaction facilitates the evolution of gene expression plasticity. BMC Biol 21, 57, DOI: 10.1186/s12915-023-01558-6. https://www.ncbi.nlm.nih.gov/pubmed/36941675. |
| Lee, C.Y., Wang, J.F., Chang, C.H., and Tung, C.W. (2023). Analyzing genomic variation in cultivated pumpkins and identification of candidate genes controlling seed traits. Plant Genome 16, e20393, DOI: 10.1002/tpg2.20393. https://www.ncbi.nlm.nih.gov/pubmed/37776006. |
| Tsai, C.H., Lai, A.C., Lin, Y.C., Chi, P.Y., Chen, Y.C., Yang, Y.H., Chen, C.H., Shen, S.Y., Hwang, T.L., Su, M.W., et al. (2023). Neutrophil extracellular trap production and CCL4L2 expression influence corticosteroid response in asthma. Sci Transl Med 15, eadf3843, DOI: 10.1126/scitranslmed.adf3843. https://www.ncbi.nlm.nih.gov/pubmed/37285400. |
2022
| Hsieh, D.K., Chuang, S.C., Chen, C.Y., Chao, Y.T., Lu, M.J., Lee, M.H., and Shih, M.C. (2022). Comparative Genomics of Three Colletotrichum scovillei Strains and Genetic Analysis Revealed Genes Involved in Fungal Growth and Virulence on Chili Pepper. Front Microbiol 13, 818291, DOI: 10.3389/fmicb.2022.818291. https://www.ncbi.nlm.nih.gov/pubmed/35154058. |
| Lu, S.H., Lee, K.Z., Hsu, P.W., Su, L.Y., Yeh, Y.C., Pan, C.Y., and Tsai, S.Y. (2022). Alternative Splicing Mediated by RNA-Binding Protein RBM24 Facilitates Cardiac Myofibrillogenesis in a Differentiation Stage-Specific Manner. Circ Res 130, 112-129, DOI: 10.1161/CIRCRESAHA.121.320080. https://www.ncbi.nlm.nih.gov/pubmed/34816743. |
| Lopez, M.L.D., Lin, Y.Y., Sato, M., Hsieh, C.H., Shiah, F.K., and Machida, R.J. (2022). Using metatranscriptomics to estimate the diversity and composition of zooplankton communities. Mol Ecol Resour 22, 638-652, DOI: 10.1111/1755-0998.13506. https://www.ncbi.nlm.nih.gov/pubmed/34555254. |
| Chiu, Y.F., Fu, H.Y., Skotnicova, P., Lin, K.M., Komenda, J., and Chu, H.A. (2022). Tandem gene amplification restores photosystem II accumulation in cytochrome b(559) mutants of cyanobacteria. New Phytol 233, 766-780, DOI: 10.1111/nph.17785. https://www.ncbi.nlm.nih.gov/pubmed/34625967. |
| Yang, F.J., Chen, C.N., Chang, T., Cheng, T.W., Chang, N.C., Kao, C.Y., Lee, C.C., Huang, Y.C., Hsu, J.C., Li, J., et al. (2022). phiC31 integrase for recombination-mediated single-copy insertion and genome manipulation in Caenorhabditis elegans. Genetics 220, DOI: 10.1093/genetics/iyab206. https://www.ncbi.nlm.nih.gov/pubmed/34791215. |
| Lu, C.W., Yao, C.T., and Hung, C.M. (2022). Domestication obscures genomic estimates of population history. Mol Ecol 31, 752-766, DOI: 10.1111/mec.16277. https://www.ncbi.nlm.nih.gov/pubmed/34779057. |
| Lu, C.W., Yao, C.T., and Hung, C.M. (2022). Domestication obscures genomic estimates of population history. Mol Ecol 31, 752-766, DOI: 10.1111/mec.16277. https://www.ncbi.nlm.nih.gov/pubmed/34779057. |
| Yeh, S.Y., Lin, H.H., Chang, Y.M., Chang, Y.L., Chang, C.K., Huang, Y.C., Ho, Y.W., Lin, C.Y., Zheng, J.Z., Jane, W.N., et al. (2022). Maize Golden2-like transcription factors boost rice chloroplast development, photosynthesis, and grain yield. Plant Physiol 188, 442-459, DOI: 10.1093/plphys/kiab511. https://www.ncbi.nlm.nih.gov/pubmed/34747472. |
| Huang, C.K., Lin, W.D., and Wu, S.H. (2022). An improved repertoire of splicing variants and their potential roles in Arabidopsis photomorphogenic development. Genome Biol 23, 50, DOI: 10.1186/s13059-022-02620-2. https://www.ncbi.nlm.nih.gov/pubmed/35139889. |
| Yang, C.L., Huang, Y.T., Schmidt, W., Klein, P., Chan, M.T., and Pan, I.C. (2022). Ethylene Response Factor109 Attunes Immunity, Photosynthesis, and Iron Homeostasis in Arabidopsis Leaves. Front Plant Sci 13, 841366, DOI: 10.3389/fpls.2022.841366. https://www.ncbi.nlm.nih.gov/pubmed/35310669. |
| Lee, T.J., Liu, Y.C., Liu, W.A., Lin, Y.F., Lee, H.H., Ke, H.M., Huang, J.P., Lu, M.J., Hsieh, C.L., Chung, K.F., et al. (2022). Extensive sampling of Saccharomyces cerevisiae in Taiwan reveals ecology and evolution of predomesticated lineages. Genome Res 32, 864-877, DOI: 10.1101/gr.276286.121. https://www.ncbi.nlm.nih.gov/pubmed/35361625. |
| Lee, T.J., Liu, Y.C., Liu, W.A., Lin, Y.F., Lee, H.H., Ke, H.M., Huang, J.P., Lu, M.J., Hsieh, C.L., Chung, K.F., et al. (2022). Extensive sampling of Saccharomyces cerevisiae in Taiwan reveals ecology and evolution of predomesticated lineages. Genome Res 32, 864-877, DOI: 10.1101/gr.276286.121. https://www.ncbi.nlm.nih.gov/pubmed/35361625. |
| Cho, H.Y., Chou, M.Y., Ho, H.Y., Chen, W.C., and Shih, M.C. (2022). Ethylene modulates translation dynamics in Arabidopsis under submergence via GCN2 and EIN2. Sci Adv 8, eabm7863, DOI: 10.1126/sciadv.abm7863. https://www.ncbi.nlm.nih.gov/pubmed/35658031. |
| Ng, C.S., Lai, C.K., Ke, H.M., Lee, H.H., Chen, C.F., Tang, P.C., Cheng, H.C., Lu, M.J., Li, W.H., and Tsai, I.J. (2022). Genome Assembly and Evolutionary Analysis of the Mandarin Duck Aix galericulata Reveal Strong Genome Conservation among Ducks. Genome Biology and Evolution 14, ARTN evac083 https://doi.org/10.1093/gbe/evac083 |
| Chang, Y.J., Kang, Z., Bei, J., Chou, S.J., Lu, M.J., Su, Y.L., Lin, S.W., Wang, H.H., Lin, S., and Chang, C.J. (2022). Generation of TRIM28 Knockout K562 Cells by CRISPR/Cas9 Genome Editing and Characterization of TRIM28-Regulated Gene Expression in Cell Proliferation and Hemoglobin Beta Subunits. Int J Mol Sci 23, DOI: 10.3390/ijms23126839. https://www.ncbi.nlm.nih.gov/pubmed/35743282. |
| Wada, N., Hsu, M.T., Tandon, K., Hsiao, S.S., Chen, H.J., Chen, Y.H., Chiang, P.W., Yu, S.P., Lu, C.Y., Chiou, Y.J., et al. (2022). High-resolution spatial and genomic characterization of coral-associated microbial aggregates in the coral Stylophora pistillata. Sci Adv 8, eabo2431, DOI: 10.1126/sciadv.abo2431. https://www.ncbi.nlm.nih.gov/pubmed/35857470. |
| Lo, K.L., Chen, Y.N., Chiang, M.Y., Chen, M.C., Panibe, J.P., Chiu, C.C., Liu, L.W., Chen, L.J., Chen, C.W., Li, W.H., and Wang, C.S. (2022). Two genomic regions of a sodium azide induced rice mutant confer broad-spectrum and durable resistance to blast disease. Rice (N Y) 15, 2, DOI: 10.1186/s12284-021-00547-z. https://www.ncbi.nlm.nih.gov/pubmed/35006368. |
| Liu, W.Y., Yu, C.P., Chang, C.K., Chen, H.J., Li, M.Y., Chen, Y.H., Shiu, S.H., Ku, M.S.B., Tu, S.L., Lu, M.J., and Li, W.H. (2022). Regulators of early maize leaf development inferred from transcriptomes of laser capture microdissection (LCM)-isolated embryonic leaf cells. Proc Natl Acad Sci U S A 119, e2208795119, DOI: 10.1073/pnas.2208795119. https://www.ncbi.nlm.nih.gov/pubmed/36001691. |
| Tsai, H.Y., Cheng, H.T., and Tsai, Y.T. (2022). Biogenesis of C. elegans spermatogenesis small RNAs is initiated by a zc3h12a-like ribonuclease. Sci Adv 8, eabm0699, DOI: 10.1126/sciadv.abm0699. https://www.ncbi.nlm.nih.gov/pubmed/35947655. |
| Chuang, C.R., Hsieh, C.L., Chang, C.S., Wang, C.M., Tandang, D.N., Gardner, E.M., Audi, L., Zerega, N.J.C., and Chung, K.F. (2022). Amis Pacilo and Yami Cipoho are not the same as the Pacific breadfruit starch crop-Target enrichment phylogenomics of a long-misidentified Artocarpus species sheds light on the northward Austronesian migration from the Philippines to Taiwan. PLoS One 17, e0272680, DOI: 10.1371/journal.pone.0272680. https://www.ncbi.nlm.nih.gov/pubmed/36178903. |
| Chou, C.H., Lin, H.S., Wen, C.H., and Tung, C.W. (2022). Patterns of genetic variation and QTLs controlling grain traits in a collection of global wheat germplasm revealed by high-quality SNP markers. Bmc Plant Biol 22, 455, DOI: 10.1186/s12870-022-03844-x. https://www.ncbi.nlm.nih.gov/pubmed/36131260. |
| Hsieh, C.L., Xu, W.B., and Chung, K.F. (2022). Plastomes of limestone karst gesneriad genera Petrocodon and Primulina, and the comparative plastid phylogenomics of Gesneriaceae. Sci Rep 12, 15800, DOI: 10.1038/s41598-022-19812-2. https://www.ncbi.nlm.nih.gov/pubmed/36138079. |
| Yeh, H.-h., Chang, Y.-M., Chang, Y.-W., Lu, M.-Y.J., Chen, Y.-H., Lee, C.-C., and Chen, C.-C. (2022). Multiomic analyses reveal enriched glycolytic processes in β-myosin heavy chain-expressed cardiomyocytes in early cardiac hypertrophy. Journal of Molecular and Cellular Cardiology Plus 1, 100011, DOI: 10.1016/j.jmccpl.2022.100011. https://www.sciencedirect.com/science/article/pii/S2772976122000058. |
2021
| Tsai, S.Y., and Huang, F. (2021). Acetyltransferase Enok regulates transposon silencing and piRNA cluster transcription. Plos Genet 17, e1009349, DOI: 10.1371/journal.pgen.1009349. https://www.ncbi.nlm.nih.gov/pubmed/33524038. |
| Lin, Y.F., Sung, C.M., Ke, H.M., Kuo, C.J., Liu, W.A., Tsai, W.S., Lin, C.Y., Cheng, H.T., Lu, M.J., Tsai, I.J., and Hsieh, S.Y. (2021). The rectal mucosal but not fecal microbiota detects subclinical ulcerative colitis. Gut Microbes 13, 1-10, DOI: 10.1080/19490976.2020.1832856. https://www.ncbi.nlm.nih.gov/pubmed/33525983. |
| Huang, C.F., Liu, W.Y., Lu, M.J., Chen, Y.H., Ku, M.S.B., and Li, W.H. (2021). Whole-Genome Duplication Facilitated the Evolution of C4 Photosynthesis in Gynandropsis gynandra. Mol Biol Evol 38, 4715-4731, DOI: 10.1093/molbev/msab200. https://www.ncbi.nlm.nih.gov/pubmed/34191030. |
| Panibe, J.P., Wang, L., Li, J., Li, M.Y., Lee, Y.C., Wang, C.S., Ku, M.S.B., Lu, M.J., and Li, W.H. (2021). Chromosomal-level genome assembly of the semi-dwarf rice Taichung Native 1, an initiator of Green Revolution. Genomics 113, 2656-2674, DOI: 10.1016/j.ygeno.2021.06.006. https://www.ncbi.nlm.nih.gov/pubmed/34111524. |
| Liu, H.H., Wang, J., Wu, P.H., Lu, M.J., Li, J.Y., Shen, Y.M., Tzeng, M.N., Kuo, C.H., Lin, Y.H., and Chang, H.X. (2021). Whole-Genome Sequence Resource of Calonectria ilicicola, the Casual Pathogen of Soybean Red Crown Rot. Mol Plant Microbe Interact 34, 848-851, DOI: 10.1094/MPMI-11-20-0315-A. https://www.ncbi.nlm.nih.gov/pubmed/33683143. |
| Li, Y.J., Wang, R.B., Lin, C.Y., Chen, S.H., Chuang, C.H., Chou, T.H., Ko, C.F., Chou, P.H., Liu, C.T., and Shih, Y.H. (2021). The degradation mechanisms of Rhodopseudomonas palustris toward hexabromocyclododecane by time-course transcriptome analysis. Chem Eng J 425, ARTN 130489 https://doi.org/10.1016/j.cej.2021.130489. |
| Lin, H.C., Wong, Y.H., Sung, C.H., and Chan, B.K.K. (2021). Histology and transcriptomic analyses of barnacles with different base materials and habitats shed lights on the duplication and chemical diversification of barnacle cement proteins. BMC Genomics 22, 783, DOI: 10.1186/s12864-021-08049-4. https://www.ncbi.nlm.nih.gov/pubmed/34724896. |
| Huang, T.H., and Suen, D.F. (2021). Iron insufficiency in floral buds impairs pollen development by disrupting tapetum function. Plant J 108, 244-267, DOI: 10.1111/tpj.15438. https://www.ncbi.nlm.nih.gov/pubmed/34310779. |
| Machida, R.J., Kurihara, H., Nakajima, R., Sakamaki, T., Lin, Y.Y., and Furusawa, K. (2021). Comparative analysis of zooplankton diversities and compositions estimated from complement DNA and genomic DNA amplicons, metatranscriptomics, and morphological identifications. Ices J Mar Sci 78, 3428-3443, DOI: 10.1093/icesjms/fsab084. https://www.webofscience.com/wos/woscc/full-record/WOS:000743803900029. |
| Huang, Y.S., Lin, C.Y., and Cheng, W.C. (2021). Investigating the Transcriptomic and Expression Presence-Absence Variation Exist in Japanese Eel (Anguilla japonica), a Primitive Teleost. Mar Biotechnol (NY) 23, 943-954, DOI: 10.1007/s10126-021-10077-w. https://www.ncbi.nlm.nih.gov/pubmed/34714446. |
| Chan, T.C., Chen, Y.T., Tan, K.T., Wu, C.L., Wu, W.J., Li, W.M., Wang, J.M., Shiue, Y.L., and Li, C.F. (2021). Biological significance of MYC and CEBPD coamplification in urothelial carcinoma: Multilayered genomic, transcriptional and posttranscriptional positive feedback loops enhance oncogenic glycolysis. Clin Transl Med 11, e674, DOI: 10.1002/ctm2.674. https://www.ncbi.nlm.nih.gov/pubmed/34954904. |
| Hsieh, C.L., Yu, C.C., Huang, Y.L., and Chung, K.F. (2021). Mahonia vs. Berberis Unloaded: Generic Delimitation and Infrafamilial Classification of Berberidaceae Based on Plastid Phylogenomics. Front Plant Sci 12, 720171, DOI: 10.3389/fpls.2021.720171. https://www.ncbi.nlm.nih.gov/pubmed/35069611. |
| Chuang, W.-H., Cheng, H.-C., Chang, Y.-J., Fu, P.-Y., Huang, Y.-C., Hsieha, P.-H., Chen, S.-H., Lina, C.-Y., and Ho, J.-M. (2021). GABOLA: A Reliable Gap-Filling Strategy for de novo Chromosome-Level Assembly. bioRxiv, 2021.2009.2007.459217, DOI: 10.1101/2021.09.07.459217. https://www.biorxiv.org/content/biorxiv/early/2021/09/08/2021.09.07.459217.full.pdf. |
| Lee, T.J., Liu, Y.-C., Liu, W.-A., Lin, Y.-F., Lee, H.-H., Ke, H.-M., Huang, J.-P., Lu, M.-Y.J., Hsieh, C.-L., Chung, K.-F., et al. (2021). Deep sampling of ancestral genetic diversity reveals Saccharomyces cerevisiae pre-domestication life histories. bioRxiv, 2021.2009.2007.459046, DOI: 10.1101/2021.09.07.459046. https://www.biorxiv.org/content/biorxiv/early/2021/09/08/2021.09.07.459046.full.pdf. |
| Fan, W.L., Yang, L.Y., Hsieh, J.C., Lin, T.C., Lu, M.J., and Liao, C.T. (2021). Prognostic Genetic Biomarkers Based on Oncogenic Signaling Pathways for Outcome Prediction in Patients with Oral Cavity Squamous Cell Carcinoma. Cancers (Basel) 13, 10.3390/cancers13112709. https://www.ncbi.nlm.nih.gov/pubmed/34070941. |
2020
| Shen, Y.C., Hsu, C.L., Jeng, Y.M., Ho, M.C., Ho, C.M., Yeh, C.P., Yeh, C.Y., Hsu, M.C., Hu, R.H., and Cheng, A.L. (2020). Reliability of a single-region sample to evaluate tumor immune microenvironment in hepatocellular carcinoma. J Hepatol 72, 489-497, DOI: 10.1016/j.jhep.2019.09.032. https://www.ncbi.nlm.nih.gov/pubmed/31634533. |
| Fontana, S., Chang, N.C., Chang, T., Lee, C.C., Dang, V.D., and Wang, J. (2020). The fire ant social supergene is characterized by extensive gene and transposable element copy number variation. Molecular Ecology 29, 105-120, DOI: 10.1111/mec.15308. https://www.webofscience.com/wos/woscc/full-record/WOS:000499403800001. |
| Wang, P.H., Chen, Y.L., Wei, S.T., Wu, K., Lee, T.H., Wu, T.Y., and Chiang, Y.R. (2020). Retroconversion of estrogens into androgens by bacteria via a cobalamin-mediated methylation. Proc Natl Acad Sci U S A 117, 1395-1403, DOI: 10.1073/pnas.1914380117. https://www.ncbi.nlm.nih.gov/pubmed/31848239. |
| Cheng, Y.I., Chou, L., Chiu, Y.F., Hsueh, H.T., Kuo, C.H., and Chu, H.A. (2020). Comparative Genomic Analysis of a Novel Strain of Taiwan Hot-Spring Cyanobacterium Thermosynechococcus sp. CL-1. Front Microbiol 11, 82, DOI: 10.3389/fmicb.2020.00082. https://www.ncbi.nlm.nih.gov/pubmed/32082292. |
| Lin, K.Y., Wang, W.D., Lin, C.H., Rastegari, E., Su, Y.H., Chang, Y.T., Liao, Y.F., Chang, Y.C., Pi, H., Yu, B.Y., et al. (2020). Piwi reduction in the aged niche eliminates germline stem cells via Toll-GSK3 signaling. Nat Commun 11, 3147, DOI: 10.1038/s41467-020-16858-6. https://www.ncbi.nlm.nih.gov/pubmed/32561720. |
| Leu, J.Y., Chang, S.L., Chao, J.C., Woods, L.C., and McDonald, M.J. (2020). Sex alters molecular evolution in diploid experimental populations of S. cerevisiae. Nat Ecol Evol 4, 453-460, DOI: 10.1038/s41559-020-1101-1. https://www.ncbi.nlm.nih.gov/pubmed/32042122. |
| Wang, W.F., Lu, M.J., Cheng, T.R., Tang, Y.C., Teng, Y.C., Hwa, T.Y., Chen, Y.H., Li, M.Y., Wu, M.H., Chuang, P.C., et al. (2020). Genomic Analysis of Mycobacterium tuberculosis Isolates and Construction of a Beijing Lineage Reference Genome. Genome Biol Evol 12, 3890-3905, DOI: 10.1093/gbe/evaa009. https://www.ncbi.nlm.nih.gov/pubmed/31971587. |
| Chang, C.Y., Hung, J.H., Huang, L.W., Li, J., Fung, K.S., Kao, C.F., and Chen, L. (2020). Epigenetic Regulation of WNT3A Enhancer during Regeneration of Injured Cortical Neurons. Int J Mol Sci 21, DOI: 10.3390/ijms21051891. https://www.ncbi.nlm.nih.gov/pubmed/32164275. |
| Vu, T.D., Iwasaki, Y., Shigenobu, S., Maruko, A., Oshima, K., Iioka, E., Huang, C.L., Abe, T., Tamaki, S., Lin, Y.W., et al. (2020). Behavioral and brain- transcriptomic synchronization between the two opponents of a fighting pair of the fish Betta splendens. Plos Genet 16, e1008831, DOI: 10.1371/journal.pgen.1008831. https://www.ncbi.nlm.nih.gov/pubmed/32555673. |
| Liu, W.Y., Lin, H.H., Yu, C.P., Chang, C.K., Chen, H.J., Lin, J.J., Lu, M.J., Tu, S.L., Shiu, S.H., Wu, S.H., et al. (2020). Maize ANT1 modulates vascular development, chloroplast development, photosynthesis, and plant growth. Proc Natl Acad Sci U S A 117, 21747-21756, DOI: 10.1073/pnas.2012245117. https://www.ncbi.nlm.nih.gov/pubmed/32817425. |
| Chen, F.Y., Jung, H.W., Tsuei, C.Y., and Liao, J.C. (2020). Converting Escherichia coli to a Synthetic Methylotroph Growing Solely on Methanol. Cell 182, 933-946 e914, DOI: 10.1016/j.cell.2020.07.010. https://www.ncbi.nlm.nih.gov/pubmed/32780992. |
| Chen, C.Y., Chen, C.K., Chen, Y.Y., Fang, A., Shaw, G.T., Hung, C.M., and Wang, D. (2020). Maternal gut microbes shape the early-life assembly of gut microbiota in passerine chicks via nests. Microbiome 8, 129, DOI: 10.1186/s40168-020-00896-9. https://www.ncbi.nlm.nih.gov/pubmed/32917256. |
| Tandon, K., Lu, C.Y., Chiang, P.W., Wada, N., Yang, S.H., Chan, Y.F., Chen, P.Y., Chang, H.Y., Chiou, Y.J., Chou, M.S., et al. (2020). Comparative genomics: Dominant coral-bacterium Endozoicomonas acroporae metabolizes dimethylsulfoniopropionate (DMSP). ISME J 14, 1290-1303, DOI: 10.1038/s41396-020-0610-x. https://www.ncbi.nlm.nih.gov/pubmed/32055028. |
| Brandon-Mong, G.J., Shaw, G.T., Chen, W.H., Chen, C.C., and Wang, D. (2020). A network approach to investigating the key microbes and stability of gut microbial communities in a mouse neuropathic pain model. BMC Microbiol 20, 295, DOI: 10.1186/s12866-020-01981-7. https://www.ncbi.nlm.nih.gov/pubmed/32998681. |
| Yang, H.C., Chen, C.H., Wang, J.H., Liao, H.C., Yang, C.T., Chen, C.W., Lin, Y.C., Kao, C.H., Lu, M.J., and Liao, J.C. (2020). Analysis of genomic distributions of SARS-CoV-2 reveals a dominant strain type with strong allelic associations. Proc Natl Acad Sci U S A 117, 30679-30686, DOI: 10.1073/pnas.2007840117. https://www.ncbi.nlm.nih.gov/pubmed/33184173. |
| Lin, C.Y., Lu, M.J., Yue, J.X., Li, K.L., Le Petillon, Y., Yong, L.W., Chen, Y.H., Tsai, F.Y., Lyu, Y.F., Chen, C.Y., et al. (2020). Molecular asymmetry in the cephalochordate embryo revealed by single-blastomere transcriptome profiling. Plos Genet 16, e1009294, DOI: 10.1371/journal.pgen.1009294. https://www.ncbi.nlm.nih.gov/pubmed/33382716. |
| Le, M.H., and Wang, D. (2020). Structure and membership of gut microbial communities in multiple fish cryptic species under potential migratory effects. Sci Rep 10, 7547, DOI: 10.1038/s41598-020-64570-8. https://www.ncbi.nlm.nih.gov/pubmed/32372020. |
| Cheng, Y.H., Liu, C.J., Yu, Y.H., Jhou, Y.T., Fujishima, M., Tsai, I.J., and Leu, J.Y. (2020). Genome plasticity in Paramecium bursaria revealed by population genomics. BMC Biol 18, 180, DOI: 10.1186/s12915-020-00912-2. https://www.ncbi.nlm.nih.gov/pubmed/33250052. |
| Liu, S.L., Chiang, Y.R., Yoon, H.S., and Fu, H.Y. (2020). Comparative Genome Analysis Reveals Cyanidiococcus gen. nov., A New Extremophilic Red Algal Genus Sister to Cyanidioschyzon (Cyanidioschyzonaceae, Rhodophyta). J Phycol 56, 1428-1442, DOI: 10.1111/jpy.13056. https://www.ncbi.nlm.nih.gov/pubmed/33460076. |
| Lee, C.H., Chang, H.W., Yang, C.T., Wali, N., Shie, J.J., and Hsueh, Y.P. (2020). Sensory cilia as the Achilles heel of nematodes when attacked by carnivorous mushrooms. P Natl Acad Sci USA 117, 6014-6022, DOI: 10.1073/pnas.1918473117. https://www.webofscience.com/wos/woscc/full-record/WOS:000520011000063. |
| Chen, W.J., Kuo, T.Y., Chen, C.Y., Hsieh, F.C., Yang, Y.L., Liu, J.R., and Shih, M.C. (2020). Whole Genome Sequencing and Tn5-Insertion Mutagenesis of Pseudomonas taiwanensis CMS to Probe Its Antagonistic Activity Against Rice Bacterial Blight Disease. Int J Mol Sci 21, DOI: 10.3390/ijms21228639. https://www.ncbi.nlm.nih.gov/pubmed/33207795. |
| Ke, H.-M., Lee, H.-H., Lin, C.-Y.I., Liu, Y.-C., Lu, M.R., Hsieh, J.-W.A., Chang, C.-C., Wu, P.-H., Lu, M.J., Li, J.-Y., et al. (2020). Mycena genomes resolve the evolution of fungal bioluminescence. Proceedings of the National Academy of Sciences 117, 31267-31277, doi:10.1073/pnas.2010761117. https://www.pnas.org/doi/abs/10.1073/pnas.2010761117. |
| An-Ting Liou, C.-H.C., Ping-Chang Lin, Yu-Chen Cheng, Ching-Yi Tsai, Wen-Chang Lin, Jeng-Yi Li, Mei-Yeh Jade Lu, Jer-Ming Chang, Shang-Jyh Hwang, Ming-Feng Hou, Inn-Wen Chong, Yuh-Jyh Jong, Jih-Jin Tsai, and Chiaho Shih (2020). GISAID: EPI_ISL_4169856 (hCoV-19/Taiwan/KMU-1/2020; B.1.438). https://epicov.org/epi3/epi_set/EPI_ISL_4169856. |
2019
| Wong, M.M., Bhaskara, G.B., Wen, T.N., Lin, W.D., Nguyen, T.T., Chong, G.L., and Verslues, P.E. (2019). Phosphoproteomics of Arabidopsis Highly ABA-Induced1 identifies AT-Hook-Like10 phosphorylation required for stress growth regulation. Proc Natl Acad Sci U S A 116, 2354-2363, DOI: 10.1073/pnas.1819971116. https://www.ncbi.nlm.nih.gov/pubmed/30670655. |
| Cho, H.Y., Lu, M.J., and Shih, M.C. (2019). The SnRK1-eIFiso4G1 signaling relay regulates the translation of specific mRNAs in Arabidopsis under submergence. New Phytol 222, 366-381, DOI: 10.1111/nph.15589. https://www.ncbi.nlm.nih.gov/pubmed/30414328. |
| Luciano, L.B.G., Tsai, I.J., Chuma, I., Tosa, Y., Chen, Y.H., Li, J.Y., Li, M.Y., Lu, M.Y.J., Nakayashiki, H., and Li, W.H. (2019). Blast Fungal Genomes Show Frequent Chromosomal Changes, Gene Gains and Losses, and Effector Gene Turnover. Mol Biol Evol 36, 1148-1161, DOI: 10.1093/molbev/msz045. https://www.webofscience.com/wos/woscc/full-record/WOS:000473587400004. |
| Lin, S.S., Chen, Y., and Lu, M.J. (2019). Degradome Sequencing in Plants. Methods Mol Biol 1932, 197-213, DOI: 10.1007/978-1-4939-9042-9_15. https://www.ncbi.nlm.nih.gov/pubmed/30701502. |
| Chang, Y.M., Lin, H.H., Liu, W.Y., Yu, C.P., Chen, H.J., Wartini, P.P., Kao, Y.Y., Wu, Y.H., Lin, J.J., Lu, M.J., et al. (2019). Comparative transcriptomics method to infer gene coexpression networks and its applications to maize and rice leaf transcriptomes. Proc Natl Acad Sci U S A 116, 3091-3099, DOI: 10.1073/pnas.1817621116. https://www.ncbi.nlm.nih.gov/pubmed/30718437. |
| Sung, C.M., Lin, Y.F., Chen, K.F., Ke, H.M., Huang, H.Y., Gong, Y.N., Tsai, W.S., You, J.F., Lu, M.J., Cheng, H.T., et al. (2019). Predicting Clinical Outcomes of Cirrhosis Patients With Hepatic Encephalopathy From the Fecal Microbiome. Cell Mol Gastroenterol Hepatol 8, 301-318 e302, DOI: 10.1016/j.jcmgh.2019.04.008. https://www.ncbi.nlm.nih.gov/pubmed/31004827. |
| Chien, C.C., Chou, M.Y., Chen, C.Y., and Shih, M.C. (2019). Analysis of genetic diversity of Xanthomonas oryzae pv. oryzae populations in Taiwan. Sci Rep 9, 316, DOI: 10.1038/s41598-018-36575-x. https://www.ncbi.nlm.nih.gov/pubmed/30670790. |
| Lin, C.C., Chao, Y.T., Chen, W.C., Ho, H.Y., Chou, M.Y., Li, Y.R., Wu, Y.L., Yang, H.A., Hsieh, H., Lin, C.S., et al. (2019). Regulatory cascade involving transcriptional and N-end rule pathways in rice under submergence. Proc Natl Acad Sci U S A 116, 3300-3309, DOI: 10.1073/pnas.1818507116. https://www.ncbi.nlm.nih.gov/pubmed/30723146. |
| Lin, C.Y.G., Chao, J.L., Tsai, H.K., Chalker, D., and Yao, M.C. (2019). Setting boundaries for genome-wide heterochromatic DNA deletions through flanking inverted repeats in Tetrahymena thermophila. Nucleic Acids Research 47, 5181-5192, DOI: 10.1093/nar/gkz209. https://www.webofscience.com/wos/woscc/full-record/WOS:000471248400028. |
| Chong, G.L., Foo, M.H., Lin, W.D., Wong, M.M., and Verslues, P.E. (2019). Highly ABA-Induced 1 (HAI1)-Interacting protein HIN1 and drought acclimation-enhanced splicing efficiency at intron retention sites. P Natl Acad Sci USA 116, 22376-22385, DOI: 10.1073/pnas.1906244116. https://www.webofscience.com/wos/woscc/full-record/WOS:000493720200064. |
| Yang, S.F., Lu, C.W., Yao, C.T., and Hung, C.M. (2019). To Trim or Not to Trim: Effects of Read Trimming on the De Novo Genome Assembly of a Widespread East Asian Passerine, the Rufous-Capped Babbler (Cyanoderma ruficeps Blyth). Genes (Basel) 10, DOI: 10.3390/genes10100737. https://www.ncbi.nlm.nih.gov/pubmed/31548511. |
| Dang, V.D., Cohanim, A.B., Fontana, S., Privman, E., and Wang, J. (2019). Has gene expression neofunctionalization in the fire ant antennae contributed to queen discrimination behavior? Ecol Evol 9, 12754-12766, DOI: 10.1002/ece3.5748. https://www.ncbi.nlm.nih.gov/pubmed/31788211. |
| Lin, Y.T., Lin, Y.F., Tsai, I.J., Chang, E.H., Jien, S.H., Lin, Y.J., and Chiu, C.Y. (2019). Structure and Diversity of Soil Bacterial Communities in Offshore Islands. Sci Rep 9, 4689, DOI: 10.1038/s41598-019-41170-9. https://www.ncbi.nlm.nih.gov/pubmed/30894580. |
2018
| Huang, C.J., Lu, M.Y., Chang, Y.W., and Li, W.H. (2018). Experimental Evolution of Yeast for High-Temperature Tolerance. Mol Biol Evol 35, 1823-1839, DOI: 10.1093/molbev/msy077. https://www.ncbi.nlm.nih.gov/pubmed/29684163. |
| Chuang, T.J., Chen, Y.J., Chen, C.Y., Mai, T.L., Wang, Y.D., Yeh, C.S., Yang, M.Y., Hsiao, Y.T., Chang, T.H., Kuo, T.C., et al. (2018). Integrative transcriptome sequencing reveals extensive alternative trans-splicing and cis-backsplicing in human cells. Nucleic Acids Res 46, 3671-3691, DOI: 10.1093/nar/gky032. https://www.ncbi.nlm.nih.gov/pubmed/29385530. |
| Chao, Y.T., Chen, W.C., Chen, C.Y., Ho, H.Y., Yeh, C.H., Kuo, Y.T., Su, C.L., Yen, S.H., Hsueh, H.Y., Yeh, J.H., et al. (2018). Chromosome‐level assembly, genetic and physical mapping of Phalaenopsis aphrodite genome provides new insights into species adaptation and resources for orchid breeding. Plant Biotechnol J 16, 2027-2041, DOI: 10.1111/pbi.12936. https://www.webofscience.com/wos/woscc/full-record/WOS:000449663300006. |
| Grillet, L., Lan, P., Li, W.F., Mokkapati, G., and Schmidt, W. (2018). IRON MAN is a ubiquitous family of peptides that control iron transport in plants. Nat Plants 4, 953-+, DOI: 10.1038/s41477-018-0266-y. https://www.webofscience.com/wos/woscc/full-record/WOS:000452308700022. |
| Wei, S.T., Wu, Y.W., Lee, T.H., Huang, Y.S., Yang, C.Y., Chen, Y.L., and Chiang, Y.R. (2018). Microbial Functional Responses to Cholesterol Catabolism in Denitrifying Sludge. mSystems 3, DOI: 10.1128/mSystems.00113-18. https://www.ncbi.nlm.nih.gov/pubmed/30417110. |
| Lee, C.Y., Hsieh, P.H., Chiang, L.M., Chattopadhyay, A., Li, K.Y., Lee, Y.F., Lu, T.P., Lai, L.C., Lin, E.C., Lee, H., et al. (2018). Whole-genome sequencing reveals unique genes that contributed to the adaptive evolution of the Mikado pheasant. Gigascience 7, DOI: 10.1093/gigascience/giy044. https://www.webofscience.com/wos/woscc/full-record/WOS:000438565400001. |
| Huang, Y.C., Dang, V.D., Chang, N.C., and Wang, J. (2018). Multiple large inversions and breakpoint rewiring of gene expression in the evolution of the fire ant social supergene. Proc Biol Sci 285, DOI: 10.1098/rspb.2018.0221. https://www.ncbi.nlm.nih.gov/pubmed/29769360. |
| Lee, C.C., and Wang, J. (2018). Rapid Expansion of a Highly Germline-Expressed Mariner Element Acquired by Horizontal Transfer in the Fire Ant Genome. Genome Biol Evol 10, 3262-3278, DOI: 10.1093/gbe/evy220. https://www.ncbi.nlm.nih.gov/pubmed/30304394. |
| Yen, Y.P., Hsieh, W.F., Tsai, Y.Y., Lu, Y.L., Liau, E.S., Hsu, H.C., Chen, Y.C., Liu, T.C., Chang, M., Li, J., et al. (2018). Dlk1-Dio3 locus-derived lncRNAs perpetuate postmitotic motor neuron cell fate and subtype identity. Elife 7, DOI: 10.7554/eLife.38080. https://www.ncbi.nlm.nih.gov/pubmed/30311912. |
| Koubkova-Yu, T.C.T., Chao, J.C., and Leu, J.Y. (2018). Heterologous Hsp90 promotes phenotypic diversity through network evolution. Plos Biol 16, ARTN e2006450 https://doi.org/10.1371/journal.pbio.2006450 |
| Qiu, B., Larsen, R.S., Chang, N.C., Wang, J., Boomsma, J.J., and Zhang, G. (2018). Towards reconstructing the ancestral brain gene-network regulating caste differentiation in ants. Nat Ecol Evol 2, 1782-1791, DOI: 10.1038/s41559-018-0689-x. https://www.ncbi.nlm.nih.gov/pubmed/30349091. |
| D, D., Hung, K.Y., and Tarn, W.Y. (2018). RBM4 Modulates Radial Migration via Alternative Splicing of Dab1 during Cortex Development. Mol Cell Biol 38, DOI: 10.1128/MCB.00007-18. https://www.ncbi.nlm.nih.gov/pubmed/29581187. |
2017
| Shaw, G.T., Liu, A.C., Weng, C.Y., Chou, C.Y., and Wang, D. (2017). Inferring microbial interactions in thermophilic and mesophilic anaerobic digestion of hog waste. PLoS One 12, e0181395, DOI: 10.1371/journal.pone.0181395. https://www.ncbi.nlm.nih.gov/pubmed/28732056. |
| Chen, C.Y., Chen, P.C., Weng, F.C., Shaw, G.T., and Wang, D. (2017). Habitat and indigenous gut microbes contribute to the plasticity of gut microbiome in oriental river prawn during rapid environmental change. PLoS One 12, e0181427, DOI: 10.1371/journal.pone.0181427. https://www.ncbi.nlm.nih.gov/pubmed/28715471. |
| Weng, F.C., Shaw, G.T., Weng, C.Y., Yang, Y.J., and Wang, D. (2017). Inferring Microbial Interactions in the Gut of the Hong Kong Whipping Frog (Polypedates megacephalus) and a Validation Using Probiotics. Front Microbiol 8, 525, DOI: 10.3389/fmicb.2017.00525. https://www.ncbi.nlm.nih.gov/pubmed/28424669. |
| Zhou, N., Swamy, K.B., Leu, J.Y., McDonald, M.J., Galafassi, S., Compagno, C., and Piskur, J. (2017). Coevolution with bacteria drives the evolution of aerobic fermentation in Lachancea kluyveri. PLoS One 12, e0173318, DOI: 10.1371/journal.pone.0173318. https://www.ncbi.nlm.nih.gov/pubmed/28282411. |
| Chao, Y.T., Yen, S.H., Yeh, J.H., Chen, W.C., and Shih, M.C. (2017). Orchidstra 2.0-A Transcriptomics Resource for the Orchid Family. Plant Cell Physiol 58, e9, DOI: 10.1093/pcp/pcw220. https://www.ncbi.nlm.nih.gov/pubmed/28111366. |
| Chung, C.L., Lee, T.J., Akiba, M., Lee, H.H., Kuo, T.H., Liu, D., Ke, H.M., Yokoi, T., Roa, M.B., Lu, M.J., et al. (2017). Comparative and population genomic landscape of Phellinus noxius: A hypervariable fungus causing root rot in trees. Mol Ecol 26, 6301-6316, DOI: 10.1111/mec.14359. https://www.ncbi.nlm.nih.gov/pubmed/28926153. |
| Chen, C.K., Yu, C.P., Li, S.C., Wu, S.M., Lu, M.J., Chen, Y.H., Chen, D.R., Ng, C.S., Ting, C.T., and Li, W.H. (2017). Identification and evolutionary analysis of long non-coding RNAs in zebra finch. BMC Genomics 18, 117, DOI: 10.1186/s12864-017-3506-z. https://www.ncbi.nlm.nih.gov/pubmed/28143393. |
| Huang, C.F., Yu, C.P., Wu, Y.H., Lu, M.J., Tu, S.L., Wu, S.H., Shiu, S.H., Ku, M.S.B., and Li, W.H. (2017). Elevated auxin biosynthesis and transport underlie high vein density in C(4) leaves. Proc Natl Acad Sci U S A 114, E6884-E6891, DOI: 10.1073/pnas.1709171114. https://www.ncbi.nlm.nih.gov/pubmed/28761000. |
| Ke, H.M., Prachumwat, A., Yu, C.P., Yang, Y.T., Promsri, S., Liu, K.F., Lo, C.F., Lu, M.J., Lai, M.C., Tsai, I.J., and Li, W.H. (2017). Comparative genomics of Vibrio campbellii strains and core species of the Vibrio Harveyi clade. Sci Rep 7, 41394, DOI: 10.1038/srep41394. https://www.ncbi.nlm.nih.gov/pubmed/28145490. |
| Merret, R., Carpentier, M.C., Favory, J.J., Picart, C., Descombin, J., Bousquet-Antonelli, C., Tillard, P., Lejay, L., Deragon, J.M., and Charng, Y.Y. (2017). Heat Shock Protein HSP101 Affects the Release of Ribosomal Protein mRNAs for Recovery after Heat Shock. Plant Physiol 174, 1216-1225, DOI: 10.1104/pp.17.00269. https://www.ncbi.nlm.nih.gov/pubmed/28381501. |
| Chen, Y.L., Yu, C.P., Lee, T.H., Goh, K.S., Chu, K.H., Wang, P.H., Ismail, W., Shih, C.J., and Chiang, Y.R. (2017). Biochemical Mechanisms and Catabolic Enzymes Involved in Bacterial Estrogen Degradation Pathways. Cell Chem Biol 24, 712-724 e717, DOI: 10.1016/j.chembiol.2017.05.012. https://www.ncbi.nlm.nih.gov/pubmed/28552583. |
| Shih, C.J., Chen, Y.L., Wang, C.H., Wei, S.T., Lin, I.T., Ismail, W.A., and Chiang, Y.R. (2017). Biochemical Mechanisms and Microorganisms Involved in Anaerobic Testosterone Metabolism in Estuarine Sediments. Front Microbiol 8, 1520, DOI: 10.3389/fmicb.2017.01520. https://www.ncbi.nlm.nih.gov/pubmed/28848528. |
| Lin, C.S., Chen, J.J.W., Chiu, C.C., Hsiao, H.C.W., Yang, C.J., Jin, X.H., Leebens-Mack, J., de Pamphilis, C.W., Huang, Y.T., Yang, L.H., et al. (2017). Concomitant loss of NDH complex-related genes within chloroplast and nuclear genomes in some orchids. Plant J 90, 994-1006, DOI: 10.1111/tpj.13525. https://www.webofscience.com/wos/woscc/full-record/WOS:000403346500013. |
2016
| Weng, F.C., Yang, Y.J., and Wang, D. (2016). Functional analysis for gut microbes of the brown tree frog (Polypedates megacephalus) in artificial hibernation. BMC Genomics 17, 1024, DOI: 10.1186/s12864-016-3318-6. https://www.ncbi.nlm.nih.gov/pubmed/28155661. |
| Tsai, S.Y., Chang, Y.L., Swamy, K.B., Chiang, R.L., and Huang, D.H. (2016). GAGA factor, a positive regulator of global gene expression, modulates transcriptional pausing and organization of upstream nucleosomes. Epigenetics Chromatin 9, 32, DOI: 10.1186/s13072-016-0082-4. https://www.ncbi.nlm.nih.gov/pubmed/27468311. |
| Lee, C.C., and Wang, J. (2016). The complete mitochondrial genome of Histiostoma blomquisti (Acari: Histiostomatidae). Mitochondrial DNA B Resour 1, 671-673, DOI: 10.1080/23802359.2016.1219633. https://www.ncbi.nlm.nih.gov/pubmed/33473592. |
| Liu, M.S., Kuo, T.C.Y., Ko, C.Y., Wu, D.C., Li, K.Y., Lin, W.J., Lin, C.P., Wang, Y.W., Schafleitner, R., Lo, H.F., et al. (2016). Genomic and transcriptomic comparison of nucleotide variations for insights into bruchid resistance of mungbean ( [L.] R. Wilczek). Bmc Plant Biol 16, ARTN 46 https://doi.org/10.1186/s12870-016-0736-1. |
| Lin, W.J., Ko, C.Y., Liu, M.S., Kuo, C.Y., Wu, D.C., Chen, C.Y., Schafleitner, R., Chen, L.F., and Lo, H.F. (2016). Transcriptomic and Proteomic Research To Explore Bruchid-Resistant Genes in Mungbean Isogenic Lines. J Agric Food Chem 64, 6648-6658, DOI: 10.1021/acs.jafc.6b03015. https://www.ncbi.nlm.nih.gov/pubmed/27508985. |
| Lin, C.Y.G., Lin, I.T., and Yao, M.C. (2016). Programmed Minichromosome Elimination as a Mechanism for Somatic Genome Reduction in Tetrahymena thermophila. Plos Genet 12, ARTN e1006403 https://doi.org/10.1371/journal.pgen.1006403 |
| Schafleitner, R., Huang, S.M., Chu, S.H., Yen, J.Y., Lin, C.Y., Yan, M.R., Krishnan, B., Liu, M.S., Lo, H.F., Chen, C.Y., et al. (2016). Identification of single nucleotide polymorphism markers associated with resistance to bruchids (Callosobruchus spp.) in wild mungbean (Vigna radiata var. sublobata) and cultivated V. radiata through genotyping by sequencing and quantitative trait locus analysis. Bmc Plant Biol 16, ARTN 159 https://doi.org/10.1186/s12870-016-0847-8 |
| Liu, T.Y., Huang, T.K., Yang, S.Y., Hong, Y.T., Huang, S.M., Wang, F.N., Chiang, S.F., Tsai, S.Y., Lu, W.C., and Chiou, T.J. (2016). Identification of plant vacuolar transporters mediating phosphate storage. Nat Commun 7, 11095, DOI: 10.1038/ncomms11095. https://www.ncbi.nlm.nih.gov/pubmed/27029856. |
| Chen, C.K., Ng, C.S., Wu, S.M., Chen, J.J., Cheng, P.L., Wu, P., Lu, M.Y.J., Chen, D.R., Chuong, C.M., Cheng, H.C., et al. (2016). Regulatory Differences in Natal Down Development between Altricial Zebra Finch and Precocial Chicken. Mol Biol Evol 33, 2030-2043, DOI: 10.1093/molbev/msw085. https://www.webofscience.com/wos/woscc/full-record/WOS:000380105900012. |
| Tsai, K.J., Lu, M.Y.J., Yang, K.J., Li, M.Y., Teng, Y.C., Chen, S., Ku, M.S.B., and Li, W.H. (2016). Assembling the Setaria italica L. Beauv. genome into nine chromosomes and insights into regions affecting growth and drought tolerance. Sci Rep-Uk 6, ARTN 35076 https://doi.org/10.1038/srep35076 |
| Yang, F.C., Chen, Y.L., Tang, S.L., Yu, C.P., Wang, P.H., Ismail, W., Wang, C.H., Ding, J.Y., Yang, C.Y., Yang, C.Y., and Chiang, Y.R. (2016). Integrated multi-omics analyses reveal the biochemical mechanisms and phylogenetic relevance of anaerobic androgen biodegradation in the environment. ISME J 10, 1967-1983, DOI: 10.1038/ismej.2015.255. https://www.ncbi.nlm.nih.gov/pubmed/26872041. |
| Chen, Y.L., Wang, C.H., Yang, F.C., Ismail, W., Wang, P.H., Shih, C.J., Wu, Y.C., and Chiang, Y.R. (2016). Identification of Comamonas testosteroni as an androgen degrader in sewage. Sci Rep-Uk 6, ARTN 35386 https://doi.org/10.1038/srep35386 |
| Feng, Y.L., Wicke, S., Li, J.W., Han, Y., Lin, C.S., Li, D.Z., Zhou, T.T., Huang, W.C., Huang, L.Q., and Jin, X.H. (2016). Lineage-Specific Reductions of Plastid Genomes in an Orchid Tribe with Partially and Fully Mycoheterotrophic Species. Genome Biol Evol 8, 2164-2175, DOI: 10.1093/gbe/evw144. https://www.ncbi.nlm.nih.gov/pubmed/27412609. |
| Huang, Y.C., Lee, C.C., Kao, C.Y., Chang, N.C., Lin, C.C., Shoemaker, D., and Wang, J. (2016). Evolution of long centromeres in fire ants. BMC Evol Biol 16, 189, DOI: 10.1186/s12862-016-0760-7. https://www.ncbi.nlm.nih.gov/pubmed/27628313. |
| Shinde, S., Villamor, J.G., Lin, W., Sharma, S., and Verslues, P.E. (2016). Proline Coordination with Fatty Acid Synthesis and Redox Metabolism of Chloroplast and Mitochondria. Plant Physiol 172, 1074-1088, DOI: 10.1104/pp.16.01097. https://www.ncbi.nlm.nih.gov/pubmed/27512016. |
| Cheng, C.Y., Young, J.M., Lin, C.G., Chao, J.L., Malik, H.S., and Yao, M.C. (2016). The piggyBac transposon-derived genes TPB1 and TPB6 mediate essential transposon-like excision during the developmental rearrangement of key genes in Tetrahymena thermophila. Genes Dev 30, 2724-2736, DOI: 10.1101/gad.290460.116. https://www.ncbi.nlm.nih.gov/pubmed/28087716. |
2015
| Hsu, H.Y., Chen, S.H., Cha, Y.R., Tsukamoto, K., Lin, C.Y., and Han, Y.S. (2015). De Novo Assembly of the Whole Transcriptome of the Wild Embryo, Preleptocephalus, Leptocephalus, and Glass Eel of Anguilla japonica and Deciphering the Digestive and Absorptive Capacities during Early Development. PLoS One 10, e0139105, DOI: 10.1371/journal.pone.0139105. https://www.ncbi.nlm.nih.gov/pubmed/26406914. |
| Tzeng, T.D., Pao, Y.Y., Chen, P.C., Weng, F.C., Jean, W.D., and Wang, D. (2015). Effects of Host Phylogeny and Habitats on Gut Microbiomes of Oriental River Prawn (Macrobrachium nipponense). PLoS One 10, e0132860, DOI: 10.1371/journal.pone.0132860. https://www.ncbi.nlm.nih.gov/pubmed/26168244. |
| Yu, C.P., Chen, S.C.C., Chang, Y.M., Liu, W.Y., Lin, H.H., Lin, J.J., Chen, H.J., Lu, Y.J., Wu, Y.H., Lu, M.Y.J., et al. (2015). Transcriptome dynamics of developing maize leaves and genomewide prediction of cis elements and their cognate transcription factors. P Natl Acad Sci USA 112, E2477-E2486, DOI: 10.1073/pnas.1500605112. https://www.webofscience.com/wos/woscc/full-record/WOS:000354390600013. |
| Chuang, Y.C., Li, W.C., Chen, C.L., Hsu, P.W., Tung, S.Y., Kuo, H.C., Schmoll, M., and Wang, T.F. (2015). Trichoderma reesei meiosis generates segmentally aneuploid progeny with higher xylanase-producing capability. Biotechnol Biofuels 8, 30, DOI: 10.1186/s13068-015-0202-6. https://www.ncbi.nlm.nih.gov/pubmed/25729429. |
| Shiao, M.S., Chang, J.M., Fan, W.L., Lu, M.Y.J., Notredame, C., Fang, S., Kondo, R., and Li, W.H. (2015). Expression Divergence of Chemosensory Genes between and Its Sibling Species and Its Implications for Host Shift. Genome Biology and Evolution 7, 2843-2858, DOI: 10.1093/gbe/evv183. https://www.webofscience.com/wos/woscc/full-record/WOS:000364951100004. |
| Lin, C.S., Chen, J.J., Huang, Y.T., Chan, M.T., Daniell, H., Chang, W.J., Hsu, C.T., Liao, D.C., Wu, F.H., Lin, S.Y., et al. (2015). The location and translocation of ndh genes of chloroplast origin in the Orchidaceae family. Sci Rep 5, 9040, DOI: 10.1038/srep09040. https://www.ncbi.nlm.nih.gov/pubmed/25761566. |
| Ruhlman, T.A., Chang, W.J., Chen, J.J., Huang, Y.T., Chan, M.T., Zhang, J., Liao, D.C., Blazier, J.C., Jin, X., Shih, M.C., et al. (2015). NDH expression marks major transitions in plant evolution and reveals coordinate intracellular gene loss. Bmc Plant Biol 15, 100, DOI: 10.1186/s12870-015-0484-7. https://www.ncbi.nlm.nih.gov/pubmed/25886915. |
| Ng, C.S., Chen, C.K., Fan, W.L., Wu, P., Wu, S.M., Chen, J.J., Lai, Y.T., Mao, C.T., Lu, M.Y., Chen, D.R., et al. (2015). Transcriptomic analyses of regenerating adult feathers in chicken. BMC Genomics 16, 756, DOI: 10.1186/s12864-015-1966-6. https://www.ncbi.nlm.nih.gov/pubmed/26445093. |
2014
| Wu, C.S., Yu, C.Y., Chuang, C.Y., Hsiao, M., Kao, C.F., Kuo, H.C., and Chuang, T.J. (2014). Integrative transcriptome sequencing identifies trans-splicing events with important roles in human embryonic stem cell pluripotency. Genome Research 24, 25-36, DOI: 10.1101/gr.159483.113. https://www.webofscience.com/wos/woscc/full-record/WOS:000329163500003. |
| Swamy, K.B., Lin, C.H., Yen, M.R., Wang, C.Y., and Wang, D. (2014). Examining the condition-specific antisense transcription in S. cerevisiae and S. paradoxus. BMC Genomics 15, 521, DOI: 10.1186/1471-2164-15-521. https://www.ncbi.nlm.nih.gov/pubmed/24965678. |
| Machida, R.J., and Lin, Y.Y. (2014). Four methods of preparing mRNA 5′ end libraries using the Illumina sequencing platform. PLoS One 9, e101812, DOI: 10.1371/journal.pone.0101812. https://www.ncbi.nlm.nih.gov/pubmed/25003736. |
| Chao, Y.T., Su, C.L., Jean, W.H., Chen, W.C., Chang, Y.C., and Shih, M.C. (2014). Identification and characterization of the microRNA transcriptome of a moth orchid Phalaenopsis aphrodite. Plant Mol Biol 84, 529-548, DOI: 10.1007/s11103-013-0150-0. https://www.ncbi.nlm.nih.gov/pubmed/24173913. |
| Sasaki, E., Zhang, X., Sun, H.G., Lu, M.Y., Liu, T.L., Ou, A., Li, J.Y., Chen, Y.H., Ealick, S.E., and Liu, H.W. (2014). Co-opting sulphur-carrier proteins from primary metabolic pathways for 2-thiosugar biosynthesis. Nature 510, 427-431, DOI: 10.1038/nature13256. https://www.ncbi.nlm.nih.gov/pubmed/24814342. |
| Lu, M.Y., Fan, W.L., Wang, W.F., Chen, T., Tang, Y.C., Chu, F.H., Chang, T.T., Wang, S.Y., Li, M.Y., Chen, Y.H., et al. (2014). Genomic and transcriptomic analyses of the medicinal fungus Antrodia cinnamomea for its metabolite biosynthesis and sexual development. Proc Natl Acad Sci U S A 111, E4743-4752, DOI: 10.1073/pnas.1417570111. https://www.ncbi.nlm.nih.gov/pubmed/25336756. |
| Ng, C.S., Wu, P., Fan, W.L., Yan, J., Chen, C.K., Lai, Y.T., Wu, S.M., Mao, C.T., Chen, J.J., Lu, M.Y., et al. (2014). Genomic organization, transcriptomic analysis, and functional characterization of avian alpha- and beta-keratins in diverse feather forms. Genome Biol Evol 6, 2258-2273, DOI: 10.1093/gbe/evu181. https://www.ncbi.nlm.nih.gov/pubmed/25152353. |
| Shih, M.C., Chou, M.L., Yue, J.J., Hsu, C.T., Chang, W.J., Ko, S.S., Liao, D.C., Huang, Y.T., Chen, J.J.W., Yuan, J.L., et al. (2014). BeMADS1 is a key to delivery MADSs into nucleus in reproductive tissues-De novo characterization of Bambusa edulis transcriptome and study of MADS genes in bamboo floral development. Bmc Plant Biol 14, Artn 179 https://doi.org/10.1186/1471-2229-14-179. |
| Chang, T.H., Lo, W.S., Ku, C., Chen, L.L., and Kuo, C.H. (2014). Molecular evolution of the substrate utilization strategies and putative virulence factors in mosquito-associated Spiroplasma species. Genome Biol Evol 6, 500-509, DOI: 10.1093/gbe/evu033. https://www.ncbi.nlm.nih.gov/pubmed/24534435. |
| Lo, W.S., Chen, H., Chen, C.Y., and Kuo, C.H. (2014). Complete Genome Sequence of Vibrio vulnificus 93U204, a Bacterium Isolated from Diseased Tilapia in Taiwan. Genome Announc 2, DOI: 10.1128/genomeA.01005-14. https://www.ncbi.nlm.nih.gov/pubmed/25278541. |
2013
| Lin, C.H., Tsai, Z.T.Y., and Wang, D.Y. (2013). Role of antisense RNAs in evolution of yeast regulatory complexity. Genomics 102, 484-490, DOI: 10.1016/j.ygeno.2013.10.008. https://www.webofscience.com/wos/woscc/full-record/WOS:000328914500008. |
| Wang, J., Wurm, Y., Nipitwattanaphon, M., Riba-Grognuz, O., Huang, Y.C., Shoemaker, D., and Keller, L. (2013). A Y-like social chromosome causes alternative colony organization in fire ants. Nature 493, 664-668, DOI: 10.1038/nature11832. https://www.ncbi.nlm.nih.gov/pubmed/23334415. |
| Su, C.L., Chao, Y.T., Yen, S.H., Chen, C.Y., Chen, W.C., Chang, Y.C., and Shih, M.C. (2013). Orchidstra: an integrated orchid functional genomics database. Plant Cell Physiol 54, e11, DOI: 10.1093/pcp/pct004. https://www.ncbi.nlm.nih.gov/pubmed/23324169. |
| Liu, W.Y., Chang, Y.M., Chen, S.C., Lu, C.H., Wu, Y.H., Lu, M.Y., Chen, D.R., Shih, A.C., Sheue, C.R., Huang, H.C., et al. (2013). Anatomical and transcriptional dynamics of maize embryonic leaves during seed germination. Proc Natl Acad Sci U S A 110, 3979-3984, DOI: 10.1073/pnas.1301009110. https://www.ncbi.nlm.nih.gov/pubmed/23431200. |
| Fan, W.L., Ng, C.S., Chen, C.F., Lu, M.Y., Chen, Y.H., Liu, C.J., Wu, S.M., Chen, C.K., Chen, J.J., Mao, C.T., et al. (2013). Genome-wide patterns of genetic variation in two domestic chickens. Genome Biol Evol 5, 1376-1392, DOI: 10.1093/gbe/evt097. https://www.ncbi.nlm.nih.gov/pubmed/23814129. |
| Shiao, M.S., Fan, W.L., Fang, S., Lu, M.Y.J., Kondo, R., and Li, W.H. (2013). Transcriptional profiling of adult antennae by high-throughput sequencing. Zool Stud 52, Artn 42 https://zoologicalstudies.springeropen.com/articles/10.1186/1810-522x-52-42. |
| Chou, M.L., Shih, M.C., Chan, M.T., Liao, S.Y., Hsu, C.T., Haung, Y.T., Chen, J.J.W., Liao, D.C., Wu, F.H., and Lin, C.S. (2013). Global transcriptome analysis and identification of a CONSTANS-like gene family in the orchid. Planta 237, 1425-1441, DOI: 10.1007/s00425-013-1850-z. https://www.webofscience.com/wos/woscc/full-record/WOS:000319474200002. |
| Schaefke, B., Emerson, J.J., Wang, T.Y., Lu, M.Y., Hsieh, L.C., and Li, W.H. (2013). Inheritance of gene expression level and selective constraints on trans- and cis-regulatory changes in yeast. Mol Biol Evol 30, 2121-2133, DOI: 10.1093/molbev/mst114. https://www.ncbi.nlm.nih.gov/pubmed/23793114. |
2012
| Chen, H.L., Chen, Y.C., Lu, M.J., Chang, J.J., Wang, H.C., Ke, H.M., Wang, T.Y., Ruan, S.K., Wang, T.Y., Hung, K.Y., et al. (2012). A highly efficient beta-glucosidase from the buffalo rumen fungus Neocallimastix patriciarum W5. Biotechnol Biofuels 5, 24, DOI: 10.1186/1754-6834-5-24. https://www.ncbi.nlm.nih.gov/pubmed/22515264. |
| Pan, I.C., Liao, D.C., Wu, F.H., Daniell, H., Singh, N.D., Chang, C., Shih, M.C., Chan, M.T., and Lin, C.S. (2012). Complete chloroplast genome sequence of an orchid model plant candidate: Erycina pusilla apply in tropical Oncidium breeding. PLoS One 7, e34738, DOI: 10.1371/journal.pone.0034738. https://www.ncbi.nlm.nih.gov/pubmed/22496851. |
| Shiao, M.S., Chang, A.Y., Liao, B.Y., Ching, Y.H., Lu, M.Y., Chen, S.M., and Li, W.H. (2012). Transcriptomes of mouse olfactory epithelium reveal sexual differences in odorant detection. Genome Biol Evol 4, 703-712, DOI: 10.1093/gbe/evs039. https://www.ncbi.nlm.nih.gov/pubmed/22511034. |
| Chang, Y.M., Liu, W.Y., Shih, A.C.C., Shen, M.N., Lu, C.H., Lu, M.Y.J., Yang, H.W., Wang, T.Y., Chen, S.C.C., Chen, S.M., et al. (2012). Characterizing Regulatory and Functional Differentiation between Maize Mesophyll and Bundle Sheath Cells by Transcriptomic Analysis. Plant Physiol 160, 165-177, DOI: 10.1104/pp.112.203810. https://www.webofscience.com/wos/woscc/full-record/WOS:000308675100017. |
2011
| Su, C.L., Chao, Y.T., Alex Chang, Y.C., Chen, W.C., Chen, C.Y., Lee, A.Y., Hwa, K.T., and Shih, M.C. (2011). De novo assembly of expressed transcripts and global analysis of the Phalaenopsis aphrodite transcriptome. Plant Cell Physiol 52, 1501-1514, DOI: 10.1093/pcp/pcr097. https://www.ncbi.nlm.nih.gov/pubmed/21771864. |
| Wang, T.Y., Chen, H.L., Lu, M.J., Chen, Y.C., Sung, H.M., Mao, C.T., Cho, H.Y., Ke, H.M., Hwa, T.Y., Ruan, S.K., et al. (2011). Functional characterization of cellulases identified from the cow rumen fungus Neocallimastix patriciarum W5 by transcriptomic and secretomic analyses. Biotechnol Biofuels 4, 24, DOI: 10.1186/1754-6834-4-24. https://www.ncbi.nlm.nih.gov/pubmed/21849025. |
2010
| Emerson, J.J., Hsieh, L.C., Sung, H.M., Wang, T.Y., Huang, C.J., Lu, H.H., Lu, M.Y., Wu, S.H., and Li, W.H. (2010). Natural selection on cis and trans regulation in yeasts. Genome Res 20, 826-836, DOI: 10.1101/gr.101576.109. https://www.ncbi.nlm.nih.gov/pubmed/20445163. |
Core News
- Single-Cell Spatial Transcriptomics Workshop
- 【 NGS 與多體學領域新知演講-2】Spatial Analysis Reveals Cancer Cell Heterogeneity: Towards Multi-Omics Spatial Analysis
- 【Survey】 Spatial Transcriptomics: Vizgen MERSCOPE Ultra and 10x Genomics Xenium Analyzer
- 【 NGS 與多體學領域新知演講】Empowering Multiomic Cytoprofiling and Next Generation Sequencing on One Platform with the AVITI24™
- We are Officially Recognized as a 10x Genomics Certified Service Provider
- 【Workshop 2024】 PacBio & BlossomBio
- 【生物資訊教育訓練課程】Partek Flow RNA/scRNA定序數據分析實作課程
- 【Workshop 2024】 Core’s tech updates and PacBio Kinnex
- NGSCore New Price Launched 服務費用更新
- 【Technology workshop 】 PacBio Kinnex Applications
- Terms of Service and Statement 服務條款與聲明
- PacBio生物資訊 教育訓練系列課程
- Technology seminar for 3D-Genomes
- 10X Genomics Technical Topics— Single cell and spatial transcriptome new knowledge and application sharing
- 【2022 Open House】Seeing the Forest through the Trees – Exploring the Cellular Diversity of Life.
- 2022 NGS Service & Introductory Workshop
- Launching of Illumina NextSeq2000 Service
- Special Discount For Users Paying by AS Intramural Funding
- Our New GridION Is Up And Running
- Spatial Transcriptome: A Novel Technology is Now Available at NGS Core
Lectures & Workshops
- 【Technology workshop 】 PacBio Kinnex Applications
- 【生物資訊教育訓練課程】臺灣 Spatial Technologies 數據分析研討會
- PacBio生物資訊 教育訓練系列課程
- 2022 NGS Service & Introductory Workshop
- 2019-10-24 Rise and Fall of Ancient Civilizations in Relation to Forest Management, and Roles of Termites in Forest Ecosystem
- 2019-05-07 PacBio Workshop: Empowering Sequencing With Highly Accurate Long Reads
- 2019-03-20 AS Core Facilities Promotion Briefings
- 2018-11-06 Core Workshop: NGS Services and Introductory
- 2018-01-24 Bionano Genomics Technology And Applications: A Solution To Produce Chromosome-scale De Novo Genome Assemblies
- 2018-01-15 10x GENOMICS Technology And Applications: Introduction of 10X Chromium System
- 2016-03-24 The Most Comprehensive View of Genomes and Transcriptomes by PacBio SMRT Sequencing
